Context
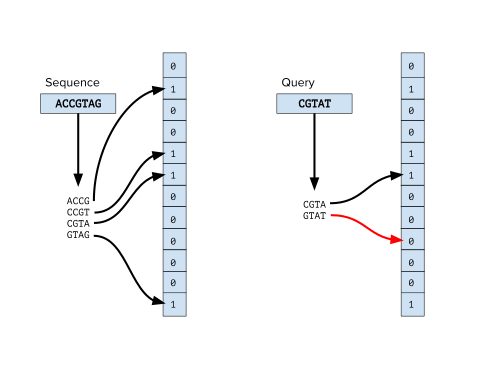
| Bloom filters are space-saving probabilistic data structures for checking whether an element is part of a set. They require much less space than other data structures to represent data sets. However, one drawback is that they can generate false positives when queried. This data structure is well used in bioinformatics to test the existence of k-mer in genomic sequences. K-mers are indexed in a Bloom filter, and any k-mer of the same size can be queried against the structure. An efficient implementation of de-Bruijn graphs relies on the use of Bloom filters.
We designed a C++ Bloom filter implementation on the UPMEM Processing-in-memory architecture. Our evaluation shows it provides interesting speedups compared to an efficient Bloom filter implementation running on CPU. For instance, inserting and querying 100 million items in a 1 GiB filter executes 2-3 times faster. |
 |
Code link
